BtuB: A Finicky Protein that Works Best at Home (DOI: https://doi.org/10.7554/eLife.68548)
By Sadie Kiendzior, UVA ChemSciComm
Have you ever wondered how bacteria uptake nutrients? I mean, bacteria have got to ‘eat’, too! A recent publication from the Cafiso group at UVA probes this exact question by closely examining a family of active protein transporters known as TonB-dependent transporters (TBDT). These proteins sit on the outer membrane (OM) of the bacterial cell and derive energy from TonB, a protein that sits between the inner and outer membranes, and interacts with inner membrane proteins ExbB and ExbD (Figure 1). Like a long game of telephone, these proteins are responsible for sensing cellular needs and initiating active transport of important molecules for survival like iron, copper, or carbohydrates into the cell. While these proteins are well understood structurally, their mechanism of transport is poorly understood. Additionally, with the core of the protein obstructed by the several amino acid residues in the core known as the core domain, as seen in Figure 2 in light yellow, there is no obvious transport path for nutrients through the protein. Thus, current hypotheses propose significant structural rearrangement of the protein, involving partial or full removal of the core domain from the surrounding barrel. The protein structure of a TBDT can be seen in Figure 2, with the outer barrel in green and core domain in light yellow. In this research, the Cafiso group probes the mechanism of action of a specific TBDT, the vitamin B12 transporter in Escherichia coli known as BtuB.
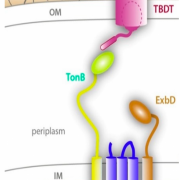
Figure 1. Schematic illustrating the location of the TonB- dependent transporter (TBDT) in relation to TonB, ExbB and ExbD. Together these proteins are responsible for actively transporting iron, carbohydrates, and other nutrients into the bacterial cell. Adapted from Celia, H. et al. Int. J. Mol. Sci. 2020, 21(2), 375; https://doi.org/10.3390/ijms21020375
Previous research has utilized a common method of purifying or partially purifying the protein and reconstituting it in a ‘model membrane’, essentially a lipid bubble, and studying protein behavior. This research suggested that upon nutrient binding, BtuB undergoes a structural change where the TonB-box unfolds and extends into the periplasm, the space between the inner and outer membrane. It is hypothesized that this event facilitates the interaction between BtuB and TonB, however no other conformational changes to the core domain of the protein were observed. Thus, in the absence of any other route for nutrient permeation, researchers hypothesize that TonB pulls the TonB-box, resulting in the unfolding of the core domain of the protein, allowing for bound molecules to pass through the core of BtuB and into the cell. An important limitation to this research is that it was all done outside of the native outer membrane environment, using purified or partially purified BtuB, which is incapable of transport.
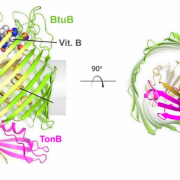
Figure 2. Structure of TBDT known as BtuB, from side and top down. Core domain is shown light yellow and outer barrel in green with TonB pictured in pink. Adapted from Celia, H., et al. Int. J. Mol. Sci. 2020, 21(2), 375; https://doi.org/10.3390/ijms21020375
A recent development in the Cafiso lab allows for unpaired electrons, also known as spin labels, to be placed on BtuB in intact E. coli cells allowing for double electron-electron resonance (DEER) measurements to be taken in fully functional BtuB. Because both of these electrons have unpaired spin states, their magnetic dipole-dipole interactions can be observed during nutrient binding to measure the change in distance between the two labels, and gather information on the dynamics of the core of the protein during transport. Preliminary studies suggested that native, or unpurified, BtuB exhibits structural changes within the core of the protein that have not been observed in purified BtuB. To probe this protein further, two electron labels were placed on BtuB, one on a fixed extracellular location and the other within the core of BtuB. Researchers found that a charge-based interaction between the core of the protein and the outer barrel was broken upon binding to transport molecules, allowing for core domain amino acid residues to extend towards the periplasmic space and subsequent transport of the nutrient into the bacterial cell. This research highlights the importance of studying proteins in their native cellular environment and revealed a mechanism of transport that was not previously hypothesized.